Pseudomonas fluorescens group bacteria as responsible for chromatic alteration on rabbit carcasses. Possible hygienic implications
Accepted: 21 February 2022
HTML: 13
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
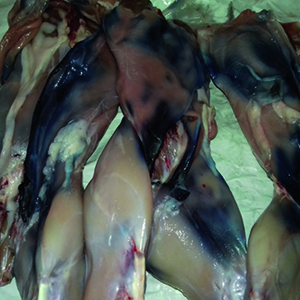
Bacteria belonging to the genus Pseudomonas are ubiquitous and characterized by a high adaptation capability to different environmental conditions and wide range of temperatures. They may colonize food, sometimes causing alteration. Quite recently, a blue pigmentation due to Pseudomonas fluorescens has been widely reported in mozzarella cheese. In this report, we describe a blue coloration occurred on rabbit meat stored in the refrigeration cell of a slaughterhouse. The alteration was observed after about 72 hours of storage at 4-6°C. Bacteriological analyses were performed, and a microorganism included in the Pseudomonas fluorescens group was identified. The experimental contamination was planned, using a bacterial suspension with 1x108 UFC/ml load to spread on rabbit carcasses. The blue pigmentation appeared after 24 hours of storage in a cell with the same conditions of temperature. The bacterium was reisolated and identified as responsible for the alteration on meat. These findings highlight the importance of considering the members of the genus Pseudomonas and, more specifically, of the P. fluorescens group when the microbiological quality of food is to be ascertained. In fact, even if these bacteria are not considered a public health problem, their presence should be monitored by food industry operators in self-control plans because they may cause alteration in food. In fact, any altered product should be withdrawn from the market in agreement with Regulation (EC) No 178/2002 of the European Parliament and of the Council.
Altinok I, Kayis S, Capkin E, 2006. Pseudomonas putida infection in rainbow trout. Aquaculture 261:850–5. DOI: https://doi.org/10.1016/j.aquaculture.2006.09.009
Andreani NA, Martino ME, Fasolato L, Carraro L, Montemurro F, Mioni R, Bordin P, Cardazzo B, 2014.Tracking the blue: A MLST approach to characterise the Pseudomonas fluorescens group. Food Microbiol 39:116–26. DOI: https://doi.org/10.1016/j.fm.2013.11.012
Angelini NM, Seigneur GN, 1988.Disease of the fins of Rhamdia sapo. Isolation of the etiological agents and experimental infection. Revista Argentina de Microbiologia 20:37–48.
Anzai Y, Kim H, Park JY, Wakabayashi H, Oyazu H, 2000. Phylogenetic affiliation of the pseudomonas based on 16S rRNA sequences. Int J Syst Evol Microbiol 50:1563–89. DOI: https://doi.org/10.1099/00207713-50-4-1563
Asghari FB, Nikaeen M, Mirhendi H, 2013. Rapid monitoring of Pseudomonas aeruginosa in hospital water system: a key priority in prevention of nosocomial infection. FEMS Microbiology Letters 343:77-81. DOI: https://doi.org/10.1111/1574-6968.12132
Bogdanova T, Flores Rodas EM, Greco S, Tolli R, Bilei S, 2010. Indagine microbiologica su campioni di mozzarella in occasione dell’allerta Mozzarella blu. In Proceedings of the XII Congresso Nazionale S.I.Di.L.V., 2010 oct 27-29, Genova, Italy, pp. 48–149.
Caldera L, Franzetti L, 2014. Effect of storage temperature on the microbial composition of ready-to-use vegetables. Curr Microbiol 68:133–9. DOI: https://doi.org/10.1007/s00284-013-0430-6
Cenci-Goga BT, Karama M, Sechi P, Iulietto MF, Novelli S, Mattei S, 2014. Evolution under different storage conditions of anomalous blue coloration of Mozzarella cheese intentionally contaminated with a pigment-producing strain of Pseudomonas fluorescens. J Dairy Sci 97:6708–18. DOI: https://doi.org/10.3168/jds.2014-8611
Circella E, Schiavone A, Barrasso R, Camarda A, Pugliese N, Bozzo G, 2020. Pseudomonas azotoformans belonging to Pseudomonas Fluorescens group as causative agent of blue coloration in carcasses of slaughterhouse rabbits. Animals 10:2-9 DOI: https://doi.org/10.3390/ani10020256
De Jonghe V, Coorevits A, Van Hoorde K, Messens W, Van Landschoot A, De Vos P, Heyndrickx M, 2011. Influence of storage conditions on the growth of Pseudomonas species in refrigerated raw milk. Appl Environ Microbiol 77:460–70. DOI: https://doi.org/10.1128/AEM.00521-10
Decimo M, Morandi S, Silvetti T, Brasca M, 2014. Characterization of Gram negative psychrotrophic bacteria isolated from Italian bulk tank milk. J Food Sci 79:2081–90. DOI: https://doi.org/10.1111/1750-3841.12645
Dominguez SA, Schaffner DW, 2007. Development and validation of a mathematical model to describe the growth of Pseudomonas spp. In raw poultry stored under aerobic conditions. Int J Food Microbiol 3:287-95. DOI: https://doi.org/10.1016/j.ijfoodmicro.2007.09.005
Drosinos EH, Board RG, 1995.Microbial and physicochemical attributes of minced lamb: Sources of contamination with pseudomonas. Food Microbiol 12:189–97. DOI: https://doi.org/10.1016/S0740-0020(95)80097-2
European Commission, 2002.Regulation of the European Parliament and of the Council of 28 January 2002 Laying down the General Principles and Requirements of Food Law, Establishing the European Food Safety Authority and Laying down Procedures in Matters of Food Safety, 178/2002/CE. In: Official Journal, L 31, 01/02/2002.
European Commission, 2004.Regulation of the European Parliament and of the Council of 29 April 2004 Laying down Specific Hygiene Rules for Food of Animal Origin, 853/2004/CE. In: Official Journal, L 139, 30/04/2004.
European Commission, 2005. Regulation of the European Parliament on Microbiological Criteria for Foodstuffs,1/2005/CE. In: Official Journal, L 338, 22/12/2005.
Evanowski RL, Reichler SJ, Kent DJ, Martin NH, Boor KJ, Wiedmann M, 2017. Pseudomonas azotoformans causes gray discoloration in HTST fluid milk. J Dairy Sci 100:7906–9. DOI: https://doi.org/10.3168/jds.2017-12650
Frapolli M, Défago G, Moënne-Loccoz Y, 2007.Multilocus sequence analysis of biocontrol fluorescent Pseudomonas spp. producing the antifungal compound 2,4-diacetylphloroglucinol. Environ Microbiol 9:1939–55. DOI: https://doi.org/10.1111/j.1462-2920.2007.01310.x
Garcia-Lopez I, Otero A, Garcia-Lopez ML, Santos JA, 2004. Molecular and phenotypic characterization of non motile Gram-negative bacteria associated with spoilage of freshwater fish. J Appl Microbiol 96:878–86. DOI: https://doi.org/10.1111/j.1365-2672.2004.02214.x
Garrido-Sanz D, Arrebola E, Martinez-Granero F, Garcia-Méndez S, Muriel C, Blanco-Romero E, Martin M, Rivilla R, Redondo-Nieto M, 2017. Classification of isolates from the Pseudomonas fluorescens complex into phylogenomic groups based in group-specific markers. Front Microbiol 8:413. DOI: https://doi.org/10.3389/fmicb.2017.00413
Gennari M, Dragotto F, 1992. A study of the incidence of different fluorescent Pseudomonas species and biovars in the microflora of fresh and spoiled meat and fish, raw milk, cheese, soil and water. J Appl Microbiol 72:281–8. DOI: https://doi.org/10.1111/j.1365-2672.1992.tb01836.x
Huang X, Madan A, 1999. CAP3: A DNA sequence assembly program. Genome Res 9:868–77. DOI: https://doi.org/10.1101/gr.9.9.868
Lane DJ, 1991. 6S/23S rRNA sequencing. In: Stackebrandt, E., Goodfellow, M., Eds. Nucleic Acid Techniques in Bacterial Systematic. John Wiley and Sons, Chichester, UK, pp. 115–175.
Mailloux RJ, Lemire J, Appanna VD, 2011. Metabolic Networks to combat oxidative stress in Pseudomonas fluorescens. Antonie van Leeuwenhoek 99:433-42. DOI: https://doi.org/10.1007/s10482-010-9538-x
Marchand S, Heylen K, Messns W, Coudijzer K, De Vos P, Dewettinck K, Herman L, De Block J, Heyndrickx M, 2009 Seasonal influence on heat-resistant proteolytic capacity of Pseudomonas lundensis and Pseudomonas fragi, predominant milk spoilers isolated from Belgian raw milk samples. Environ Microbiol 11:467–82. DOI: https://doi.org/10.1111/j.1462-2920.2008.01785.x
Martin NH, Murphy SC, Ralyea RD, Wiedmann M, Boor KJ, 2011. When cheese gets the blues: Pseudomonas fluorescens as the causative agent of cheese spoilage. J Dairy Sci 94:3176–83. DOI: https://doi.org/10.3168/jds.2011-4312
Mena KD, Gerba CP, 2009. Risk assessment of Pseudomonas aeruginosa in water. In: Review of Environmental contamination and Toxicology. Springer, New York, NY, pp 71-115. DOI: https://doi.org/10.1007/978-1-4419-0032-6_3
Nogarol C, Acutis PL, Bianchi DM, Maurella C, Peletto S, Gallina S, Adriano D, Zuccon F, Borrello S, Caramelli M, Decastelli L, 2013. Molecular characterization of Pseudomonas fluorescens isolates involved in the Italian “blue mozzarella” event. J Food Prot 76:500–504. DOI: https://doi.org/10.4315/0362-028X.JFP-12-312
Palleroni NJ, Genus I, 2005. Pseudomonas Migula 1894, 237AL (Nom. Cons., Opin. 5 of the Jud. Comm. 1952). In: Brenner DJ, Krieg NR, Staley JT, Garrity GM, Eds. Bergey’s Manual of Systematic Bacteriology, 2nd ed. Springer: New York, NY, USA, Vol 2, Part B, pp 328–379.
Peix A, Ramirez-Bahena MH, Velazquez E, 2009.Historical evolution and current status of the taxonomy of genus Pseudomonas. Infect Genet Evol 9:1132–47. DOI: https://doi.org/10.1016/j.meegid.2009.08.001
Rossi C, Serio A, Chaves-Lòpez C, Annibali F, Auricchio B, Goffredo E, Cenci-Goga BT, Lista F, Fillo S, Paparella A, 2018. Biofilm formation, pigment production and motility in Pseudomonas spp. isolated from dairy industry. Food Control 86:241–8. DOI: https://doi.org/10.1016/j.foodcont.2017.11.018
Schwarz G, Bauder R, Speer M, Rommel TO, Lingens F, 1989. Microbial metabolism of quinoline and related compounds. Degradation of quinoline by Pseudomonas fluorescens 3, Pseudomonas putida 86 and Rhodococcus spec B1. Biol Chem Hoppe-Seyler 370:1183-89. DOI: https://doi.org/10.1515/bchm3.1989.370.2.1183
Tassew H, Abdissa A, Beyene G, Gebre-Selassie S, 2011. Microbial flora and food borne pathogens on minced meat and their susceptibility to antimicrobial agents. Ethiop J Health Sci 20:137-43. DOI: https://doi.org/10.4314/ejhs.v20i3.69442
Tryfinopoulou P, Drosinos EH, Nychas GJ, 2001. Performance of Pseudomonas CFC-selective medium in the fish storage ecosystem. J Microbiol Methods 47:243-7. DOI: https://doi.org/10.1016/S0167-7012(01)00313-X
Tümmler B, Wiehlmann L, Klockgether J, Cramer N, 2014.Advances in understanding Pseudomonas. F1000 Prime Rep 6:9. DOI: https://doi.org/10.12703/P6-9
Zeng X, Chen X, Zhang W, 2015. Characterization of the microbial flora from suan yu, a chinese traditional low salt fermented fish. J Food Process Preserv 40:1093-3. DOI: https://doi.org/10.1111/jfpp.12690
PAGEPress has chosen to apply the Creative Commons Attribution NonCommercial 4.0 International License (CC BY-NC 4.0) to all manuscripts to be published.


 https://doi.org/10.4081/ijfs.2022.9998
https://doi.org/10.4081/ijfs.2022.9998



