Retrotransposon mapping in spider monkey genomes of the family Atelidae (Platyrrhini, Primates) shows a high level of LINE-1 amplification
Accepted: October 2, 2022
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
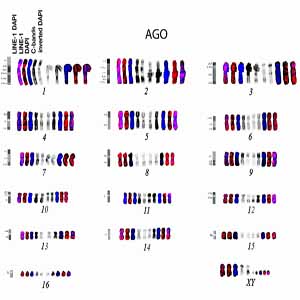
To investigate the distribution of LINE-1 repeat sequences, a LINE-1 probe was Fluorescence In Situ Hybridized (FISH) on the chromosomes of Ateles geoffroyi and Ateles fusciceps (Atelidae); a LINE-1 probe was also mapped on Cebuella pygmaea (Cebidae) and used as an outgroup for phylogenetic comparison. Ateles spider monkeys have a highly rearranged genome and are an ideal model for testing whether LINE-1 is involved in genome evolution. The LINE-1 probe has been mapped in the two Atelidae species for the first time, revealing a high accumulation of LINE-1 sequences along chromosomal arms, including telomeres, and a scarcity of LINE-1 signals at centromere positions. LINE-1 mapping in C. pygmaea (Cebidae) revealed signals at centromere positions and along chromosome arms, which was consistent with previous published data from other Cebidae species. In a broader sense, the results were analyzed in light of published data on whole-chromosomal human probes mapped in these genomes. This analysis allows us to speculate about the presence of LINE-1 sequences at the junction of human chromosomal syntenies, as well as a possible link between these sequences and chromosomal rearrangements.
Ahmad SF, Singchat W, Jehangir M, et al. Dark matter of primate genomes: Satellite DNA repeats and their evolutionary dynamics. Cells 2020:9:2714.
Mazzoleni S, Rovatsos M, Schillaci O, Dumas F. Evolutionary insight on localization of 18S, 28S rDNA genes on homologous chromosomes in primates genomes. Comp Cytogenet 2018;12:27–40.
Mazzoleni S, Schillaci O, Sineo L, Dumas F. Distribution of interstitial telomeric sequences in primates and the pygmy tree shrew (Scandentia). Cytogenet Genome Res 2017;151:141–50.
Ceraulo S, Perelman PL, Mazzoleni S, et al. Repetitive sequence distribution on Saguinus, Leontocebus and Leontopithecus Tamarins (Platyrrhine, Primates) by mapping telomeric (TTAGGG) motifs and rDNA loci. Biology 2021;10:844.
Böhne A, Brunet F, Galiana-Arnoux D, et al. Transposable elements as drivers of genomic and biological diversity in vertebrates. Chromosome Res 2008;16:203–15.
Paço A, Adega F, Chaves R. Line-1 retrotransposons: From “parasite” sequences to functional elements. J Appl Genet 2015;56:133–45.
Paço A, Freitas R, Vieira-da-Silva A. Conversion of DNA sequences: From a transposable element to a tandem repeat or to a gene. Genes 2016;10:1014.
Gray YH. It takes two transposons to tango: transposable element-mediated chromosomal rearrangements. Trends Genet 2000;16:461–68.
Bulazel KV, Ferreri GC, Eldridge MD, O'Neill RJ. Species specific shifts in centromere sequence composition are coincident with breakpoint reuse in karyotypically divergent lineages. Genome Biol 2007;8:1.
Belyayev A. Bursts of transposable elements as an evolutionary driving force. J Evol Biol 2014;27:2573–84.
Boissinot S, Furano AV. Adaptive evolution in LINE-1 retrotransposons. Mol Biol Evol 2001;18:2186–94.
Klein SJ, O’Neill RJ. Transposable elements: genome innovation, chromosome diversity, and centromere conflict. Chromosome Res 2018;26:5-23.
Richardson SR, Doucet AJ, Kopera HC, et al. The influence of LINE-1 and SINE retrotransposons on mammalian genomes. Microbiol Spectr 2015;3:1165–208.
Ovchinnikov I, Rubin A, Swergold GD. Tracing the LINEs of human evolution. Proc Natl Acad Sci USA 2002;6;99:10522-7.
Tang W, Mun S, Joshi A, et al. Mobile elements contribute to the uniqueness of human genome with 15,000 human-specific insertions and 14 Mbp sequence increase. DNA Res 2018;25:521-33.
Mathews LM, Chi SY, Greenberg N, et al. Large differences between LINE-1 amplification rates in the human and chimpanzee lineages. Am J Hum Genet 2003;72:739-48.
Kim YJ, Han K. Endogenous retrovirus-mediated genomic variations in chimpanzees. Mob Genet Elements 2015;4:1–4.
Seuánez HN, Forman L, Matayoshi T, Fanning TG. The Callimico goeldii (Primates, Platyrrhini) genome: karyology and middle repetitive (LINE- 1) DNA sequences. Chromosoma 1989;98:389–95.
Boissinot S, Roos C, Furano AV. Different rates of LINE-1 (L1) retrotransposon amplification and evolution in New World monkeys. J Mol Evol 2004;58:122-30.
Sookdeo A, Ruiz-García M, Schneider H, Boissinot S. Contrasting rates of LINE-1 amplification among New World Primates of the Atelidae family. Cytogenetic Genome Res 2018;154:217-28.
Tang W, Liang P. Comparative genomics analysis reveals high levels of differential retrotransposition among primates from the hominidae and the cercopithecidae families. Genome Biol Evol 2019;11:3309–25.
Lee S, Tang W, Liang P, Han K. A comprehensive analysis of chimpanzee (Pan troglodytes)-specific LINE-1 retrotransposons. Gene 2019;693:46-51.
Jeon S, Kim S, Oh MH, et al. A comprehensive analysis of gorilla-specific LINE-1 retrotransposons. Genes & Genomics 2021;43:1133-41.
Waters PD, Dobigny G, Pardini AT, Robinson TJ. LINE-1 distribution in Afrotheria and Xenarthra: implications for understanding the evolution of LINE-1 in eutherian genomes. Chromosoma 2004;113:137–44.
Ovchinnikov I, Troxel AB, Swergold GD. Genomic characterization of recent human LINE-1 insertions: evidence supporting random insertion. Genome Res 2001;11:2050–58.
Parish D, Vise P, Wichman H, et al. Distribution of LINEs and other repetitive elements in the karyotype of the bat Carollia: implications for X-chromosome inactivation. Cytogenet Genome Res 2002;96:191–97.
Kapitonov VV, Holmquist GP, Jurka J. L1 repeat is a basic unit of heterochromatin satellites in cetaceans. Mol Biol Evol 1998;15:611–12.
Acosta MJ, Marchal JA, Fernández-Espartero CH. Retroelements (LINEs and SINEs) in vole genomes: differential distribution in the constitutive heterochromatin. Chromosome Res 2008;16:949–59.
Bulazel K, Metcalfe C, Ferreri GC, et al. Cytogenetic and molecular evaluation of centromere-associated DNA sequences from a marsupial (Macropodidae: Macropus rufogriseus) X chromosome. Genetics 2006;172:1129–37.
Carbone L, Harris RA, Mootnick AR, et al. Centromere remodelling in Hoolock leuconedys (Hylobatidae) by a new transposable element unique to the gibbons. Genome Biol Evol 2012;4:760–70.
De Sotero-Caio CG, Cabral-de-Mello DC, da Silva Calixto M, et al. Centromeric enrichment of LINE-1 retrotransposons and its significance for the chromosome evolution of Phyllostomid bats. Chromosome Res 2017;25:313-25.
Rebuzzini P, Castiglia R, Nergadze SG, et al. Quantitative variation of LINE-1 sequences in five species and three subspecies of the subgenus Mus and in five Robertsonian races of Mus musculus domesticus. Chromosome Res 2009;17:65–76.
Perelman P, Johnson WE, Roos C, et al. A molecular phylogeny of living primates. PLoS Genetics 2011;73:e1001342.
Dumas F, Sineo L. The evolution of human synteny 4 by mapping sub-chromosomal specific probes in Primates. Caryologia 2014;67:281–91.
Dumas F, Sineo L, Ishida T. Taxonomic identification of Aotus (Platyrrhinae) through cytogenetics. [Identificazione tassonomica di Aotus (Platyrrhinae) mediante la citogenetica.] J Biol Res 2015;88:65-6.
Serfaty DMB, Carvalho NDM, Gross MC, et al.Differential chromosomal organisation between Saguinus midas and Saguinus bicolor with accumulation of differences in the repetitive sequence DNA. Genetica 2017;145:359-69.
Ceraulo S, Perelman PL, Dumas F. Massive LINE-1 retrotransposon enrichment in tamarins of the Cebidae family (Platyrrhini, Primates) and its significance for genome evolution. J Zoolog System Evolut Res 2021;59:2553–61.
Milioto V, Perelman PL, La Paglia L, et al. Mapping retrotransposonLINE-1 sequences into two cebidae species and Homo sapiens genomes and a short review on primates. Genes 2022;13:1742.
Ceraulo S, Milioto V, Dumas F. Centromeric enrichment of LINE-1 retrotransposon in two species of South American monkeys Alouatta belzebul and Ateles nancymaae (Platyrrhini, Primates). Caryologia 2021;74:111-19.
Neusser M, Stanyon R, Bigoni F, et al. Molecular cytotaxonomy of New World monkeys (Platyrrhini) – comparative analysis of five species by multi-color chromosome painting gives evidence for a classification of Callimico goeldii within the family of Callitrichidae. Cytogenet Cell Genet 2001;94:206–15.
Morescalchi MA, Schempp W, Wienberg J, Stanyon R. Chromosome painting in the New World monkey, Ateles geoffroyi, the black-handed spider monkey. Chromosome Res 1997;5:527–36.
Garcia F, Ruiz-Herrera A, Egozcue J, et al. Chromosomal homologies between Cebus and Ateles (Primates) based on ZOO-FISH and G-banding comparisons. Am J Primatol 2002;57:177-88.
De Oliveira EHC, Neusser DM, Pieczarka JC, et al. Phylogenetic inferences of Atelinae (Platyrrhini) based on multi-directional chromosome painting in Brachyteles arachnoides, Ateles paniscus paniscus and Ateles b. marginatus. Cytog Genome Res 2005;108:183-90.
Nagamachi CY, Pieczarka JC, de Souza Barros RM. Karyotypic comparison among Cebuella pygmaea, Callithrix jacchus and C. emiliae (Callitrichidae, Primates) and its taxonomic implications. Genetica 1992;85:249-57.
Fernàndez R, Barragàn M, Bullejos M, et al. New C-band protocol by heat denaturation in the presence of formamide. Hereditas 2002;137:145-8.
Nieves M, Ascunce MS, Rahn MI, Mudry MD. Phylogenetic relationships among some Ateles species: the use of chromosomal and molecular characters. Primates 2005;46:155-64.
Fantini L, Jeffery NW, Pierossi P, et al. Qualitative and quantitative analysis of the genomes and chromosomes of spider monkeys (Primates: Atelidae). Biol J Linnean Soc 2016;118:752-62.
Medeiros MA, Barros RMS, Pieczarka JC, et al. Radiation and speciation of spider monkeys, genus Ateles, from the cytogenetic viewpoint. Am J Primatol 1997;42:167-78.
Scardino R, Milioto V, Proskuryakova AA, et al. Evolution of the human chromosome 13 synteny: Evolutionary rearrangements, plasticity, human disease genes and cancer breakpoints. Genes 2020;11:383.
Scardino R, Mazzoleni S, Rovatsos M, et al. Molecular cytogenetic characterization of the sicilian endemic pond turtle Emys trinacris and the yellow-bellied slider Trachemys scripta scripta (Testudines, Emydidae). Genes 2020;11:702.
Milioto V, Vlah S, Mazzoleni S, et al. Chromosomal localization of 18S-28S rDNA and (TTAGGG)n Sequences in two south african dormice of the genus Graphiurus (Rodentia: Gliridae). Cytogenet Genome Res 2019;158:145-51.
Lemskaya NA, Kulemzina AI, Beklemisheva VR, et al. A combined banding method that allows the reliable identification of chromosomes as well as differentiation of AT-and GC-rich heterochromatin. Chromosome Res 2018;26:307-15.
Dumas F, Mazzoleni S. Neotropical primate evolution and phylogenetic reconstruction using chromosomal data. Eur Zool J 2017;84:1-18.
Cellamare A, Catacchio CR, Alkan C, et al. New insights into centromere organization and evolution from the white-cheeked gibbon and marmoset. Molec Biol Evol 2009;26:1889-900.
Logsdon GA, Vollger MR, Hsieh P, et al. The structure, function and evolution of a complete human chromosome 8. Nature 2021;593:101-7.
Nurk S, Koren S, Rhie A, et al.The complete sequence of a human genome. Science 2022;376:44-53.
Copyright (c) 2022 the Authors

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
PAGEPress has chosen to apply the Creative Commons Attribution NonCommercial 4.0 International License (CC BY-NC 4.0) to all manuscripts to be published.


 https://doi.org/10.4081/jbr.2022.10725
https://doi.org/10.4081/jbr.2022.10725



